Nader Aryamanesh
PhD, Molecular Biology and Bioinformatics
Dr Nader Aryamanesh the Bioinformatics Facility Lead at CMRI and Conjoint Senior Lecturer at the University of Sydney. He is experienced in multiomics analyses, collaborating with several research groups within CMRI as well as other medical research institutions such as Kids Research Institute and Garvan Institute of Medical Research. Before joining CMRI in 2020, Dr Aryamanesh worked as a Senior Bioinformatician/Computational Biologist in several research institutions including i) ARC Centre of Excellence in Plant Energy Biology, UWA, Australia, ii) Biocenter Oulu, University of Oulu, Finland, and iii) Bioinformatics Core, Precision Medicine Theme, South Australian Health and Medical Research Institute (SAHMRI), Adelaide, Australia. Dr Aryamanesh is interested in precision medicine through the identification of biomarkers for different human diseases including cancer and childhood disease, understanding the underlying pathways involved in diseases, stem cell research, embryogenesis, and development of novel NGS sequencing technologies.
Dr Aryamanesh is collaborating on several single cell multiomics projects (scRNA-Seq and scATAC-Seq) related to embryology, stem cell research, iPSc, organoids, eye-, liver-, adrenal gland-related diseases, and genome stability. Dr Aryamanesh has used several public scRNA-Seq databases to create Atlas databases for annotation of cell types, cell differentiation, Pseudotime trajectory and the identification of progenitor cells. Dr Aryamanesh published a workflow as a book chapter outlining the procedure for analysing scRNA-Seq data along with annotation with an Atlas reference database (Aryamanesh 2022).

Figure 1. Creation of Atlas database for adrenal gland. A) UMAP recreated with different algorithm to normalize and cluster the data from published public database Lopez et. Al. (2021). B) Original UMAP from Lopez et. Al. (2021) Science Advances, 7(5).
Dr Aryamanesh has extensive experience in handling bulk RNA-Seq, miRNA-Seq data and functional genomics. He has developed reproducible pipelines to process the transcriptomics data and perform various downstream analyses such as GO enrichment analysis, pathway analysis, copy number variants (CNV), structural variations (SVs), alternative splicing, gene fusions and footprints analysis.

Figure 2. RNA-Seq analysis of putative “footprints” in the rps12 intron halves in WT (blue) and mutant (orange). Source: Aryamanesh et. al. 2017
Dr Aryamanesh has been working on targeted sequencing (WES), Illumina WGS and long-read ONT WGS data. The analysis incudes mapping, germline variant calling, somatic variant calling, annotating the pathogenic variants using different tools (e.g., SnpEff, Funcotator, Varsome). Dr Aryamanesh is also interested in telomere length and variant repeat measurements. Dr Aryamanesh is collaborating with other researchers in CMRI and Garvan Institute of Medical Research on the effect the telomere structural variants and length in drug resistant ER+ breast cancer.
Dr Aryamanesh is also an interested in developing new sequencing technologies. He has a patent (US-2020370093-A1) entitled “methods for preparing RNA probes for exome sequencing and for depleting organelle DNA”. He is also interested in developing a technology for sequencing human telomeres.
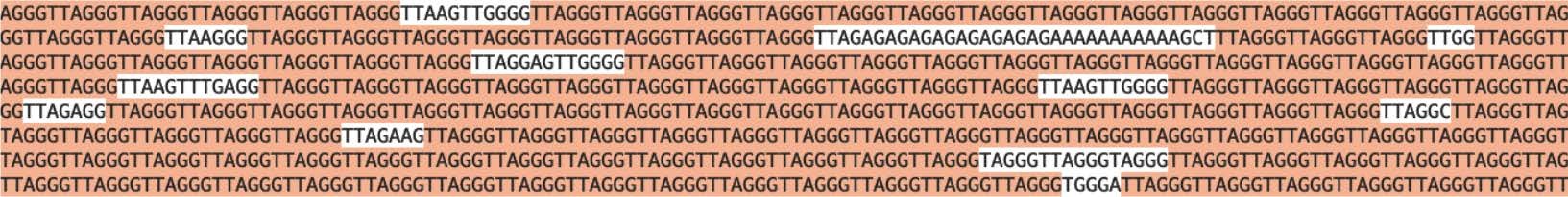
Figure 3. A screenshot of telomeric repeat from a long-read ONT sequencing showing TTAGGG repeats along with some mutations in the sequence.
Dr Aryamanesh has worked on data analyses of variety of technologies related to epigenetics including whole genome bisulfite sequencing (WGBS), EPIC Array, ATAC-Seq, ChIP-Seq, Cut&Run Assay and DamID-Seq. Most of the datasets comes with transcriptomics and genomics to dissect the underlying effect of methylation on gene expression patterns on specific disease/condition. Dr Aryamanesh is using multiomics approach to identify signature biomarkers for underlying condition.

Figure 4. Illustration of DamID-Seq identifying the binding sites of LHX1 protein in mouse neuruloids and embryos. Source: McMahon R. (2022) “A Mechanistic Insight into Building the Embryonic Head”, PhD Thesis, The University of Sydney.
Aryamanesh N. (2022). A Reproducible and Dynamic Workflow for Analysis and Annotation of scRNA-Seq Data. Methods in molecular biology (Clifton, N.J.), 2490, 101–140. https://doi.org/10.1007/978-1-0716-2281-0_10
McMahon, R., Sibbritt, T., Aryamanesh, N., Masamsetti, V. P., & Tam, P. (2022). Loss of Foxd4 Impacts Neurulation and Cranial Neural Crest Specification During Early Head Development. Frontiers in cell and developmental biology, 9, 777652. https://doi.org/10.3389/fcell.2021.777652
Okada, T., McIlfatrick, S., Hin, N., Aryamanesh, N., Breen, J., & St John, J. C. (2022). Mitochondrial supplementation of Sus scrofa metaphase II oocytes alters DNA methylation and gene expression profiles of blastocysts. Epigenetics & chromatin, 15(1), 12. https://doi.org/10.1186/s13072-022-00442-x
Aryamanesh, N., Ruwe, H., Sanglard, L. V., Eshraghi, L., Bussell, J. D., Howell, K. A., Small, I., & des Francs-Small, C. C. (2017). The Pentatricopeptide Repeat Protein EMB2654 Is Essential for Trans-Splicing of a Chloroplast Small Ribosomal Subunit Transcript. Plant physiology, 173(2), 1164–1176. https://doi.org/10.1104/pp.16.01840
Hooper, C. M., Castleden, I. R., Tanz, S. K., Aryamanesh, N., & Millar, A. H. (2017). SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Research, 45(D1), D1064–D1074. https://doi.org/10.1093/nar/gkw1041